Cleaning up PCR Samples
Evaluating Pros and Cons of Reagents Applied in Food Testing Research Applications
Two of the biggest challenges for a diagnostic laboratory using Polymerase Chain Reaction (PCR) technology are cross-contamination of PCR amplicon between samples, leading to false-positive results, and positive PCR results arising from the detection of DNA from dead cells when only the presence of live organisms is of interest or concern. For the food testing laboratory, the occurrence of false-positive results will have a negative cost impact, on both the laboratory and the wider business, due to having to repeat tests and delays in reporting results. The impact may be even greater when a false positive result goes undetected, leading to unnecessary product diversion or disposal.
To reduce these occurrences of false positives, researchers commonly include particular reagents when running PCR reactions. While this practice has been shown to be useful and effective in research laboratory circumstances, the use of these reagents in PCR assays for the detection of microbial targets in food matrices is not well documented. Thermo Fisher Scientific has now undertaken some in-house studies to ascertain the potential benefits of incorporating specific agents into PCR assays used for the detection of pathogens in foods and associated samples.
Prevention of Cross-Contamination with PCR Amplicon Using Uracil-DNA Glycosylase (UDG) and Deoxyuridine Triphosphate (dUTP)
When preparing PCR reactions it is important to separate pre- and post-PCR areas to avoid contamination of the sample with PCR products from previous runs. If such contamination should happen, it can be removed by treating the sample prior to PCR with UDG to cleave the PCR products. UDG is an enzyme that removes any uracil incorporated into single- or double-stranded DNA which may be present as a contaminant from a previous sample. The treatment of sample preparations with UDG prior to thermal cycling in combination with the use of dUTP in the PCR amplification is commonly used to prevent cross-contamination. If being used, both dUTP and dTTP (deoxythymine triphosphate) are generally incorporated in the master mix or PCR pellet of a PCR assay for this purpose.
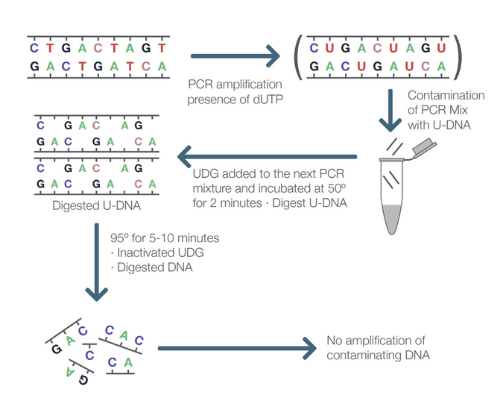
However, there are some drawbacks when using UDG:
- Usually the PCR reaction contains both dUTP and dTTP in a 50:50 or 25:75 ratio. Higher dUTP ratios have been noted to lower the efficiency of the reaction, reducing the number of copies of the target. This can lead to a false-negative result due to the target not reaching sufficient numbers to cross the cycle threshold (Ct).
- Since most PCR reactions contain both dUTP and dTTP, some of the produced PCR products do not contain dUTP bases. Most polymerases favor the incorporation of dTTP instead of dUTP if both nucleotides are present in the reaction. This leads to a high probability that some of the resulting PCR products do not contain any U bases, and therefore will not be cleaved by UDG and possible contaminants will not be removed.
- The UDG treatment works best with thymine rich amplicons but has reduced activity when amplicons are G-C rich, again increasing the possibility of incomplete contaminant removal which in turn can lead to a false-positive result.
- When the UDG treatment is used, the PCR reaction is held at 50°C before actual PCR. At this temperature, primers can form dimers and if the DNA polymerase is active, the primer-dimers will be polymerized to short DNA products. If these short DNA products are present in the PCR reaction, the polymerase will replicate them along with the desired PCR product throughout the whole PCR protocol. The probe only detects the correct PCR product, so the shorter fragments generated from primer-dimers are not detected. Instead, they will compete with the target for the components in the reaction mix required for amplification and therefore potentially reduce the sensitivity. Even the use of hot-start polymerases have been seen to produce primer-dimers due to the incubation of UDG at 50°C prior to starting the PCR cycle.
- UDG may not be completely inactivated and the residual enzymatic activity may be enough to degrade amplification products generated during the early amplification cycles resulting in delayed Ct values and lower sensitivity.
When considering the disadvantages mentioned above and the additional cost of the UDG enzyme, there may not be sufficient justification for the routine use and incorporation of the UDG enzyme in PCR assays.
Use of Photoreactive Intercalating Dyes for Prevention of Dead cell DNA Amplification and Detection by PCR
It is possible to hinder the amplification of DNA from dead cells during PCR by carrying out a pre-treatment with a photoreactive intercalating dye such as propidium monoazide (PMA) and ethidium monoazide (EMA). This approach is referred to as viability PCR (vPCR). The DNA intercalating agents are able to enter into dead bacteria with compromised cell membranes and covalently bind to DNA when exposed to strong visible light. The amplification of the modified dead-cell DNA is disturbed in PCR, while the DNA from live cells remains untouched and available for amplification. Once the dye is intercalated with the DNA of a dead or metabolically inactive cell, photoirradiation produces a highly reactive nitrene radical that reacts with any organic molecule in proximity, such as DNA. The reaction occurs due to the dye reacting with any hydrocarbon moiety, forming an irreversible covalent nitrogen-carbon bond. Any free remaining reagent reacts with water molecules upon light exposure forming a hydroxylamine that is unable to form subsequent covalent bonds and so will not go on to affect any live cells present in the sample during later stages of the assay. This prevents the amplification of DNA extracted from nonviable cells after the dye treatment.
The use of these dyes would seem to provide the answer to differentiate between live and dead cells when using PCR technology for detection of pathogens in food samples, and aid in reducing the number of PCR positive, culture confirmation negative results.
However, the dyes are not 100% specific or sensitive, and complete signal suppression is not always achieved.
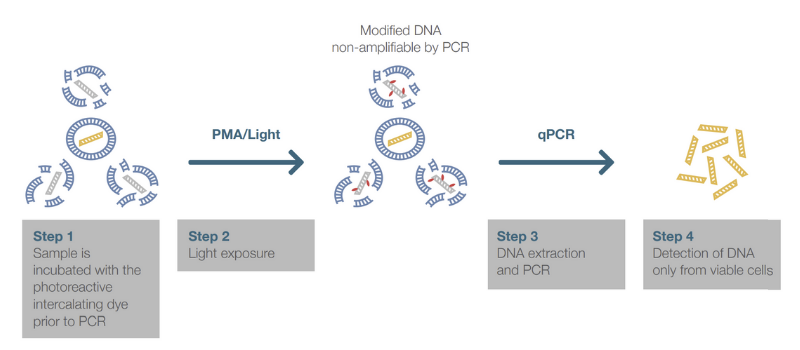
Factors Affecting the use of Photoreactive Intercalating Dyes
With more in-depth work using these dyes we have found them to have considerable limitations when working with food samples, as demonstrated by the following effects:
Choice of dye
EMA has been reported to have the ability to intercalate into the DNA of live cells resulting in a false signal suppression from target organisms, potentially leading to false-negative results; PMA has a very low affinity for live cells but some affinity for minimally injured cells, meaning that while it does not suppress detection of live cells it can be active against cells that have their membrane compromised but are otherwise viable, and in some cases lead to them not being detected. However, further investigations to understand the impact of using these dyes as a method of vPCR and viable-but-non-culturable cells remain to be investigated.
Incubation duration
Light exposure should last long enough for DNA intercalation to occur. However, excess exposure could damage the DNA of the live target organism. The temperature used should favor the intercalation of the dye into the DNA but should not be so high that it lyses live cells due to excess heating, leading to false-negative results.
Presence of suspended particles
The effectiveness of the light exposure step is affected by the turbidity of the sample. Samples that are not optically clear may lead to inefficient performance of the dyes, providing false-positive results. This is because the dyes have not been adequately inactivated, leading to DNA from dead cells being amplified and detected during PCR. This can be a problem for highly fatty foods or matrices with high background numbers such as whole chicken or ground products. It is also possible that excessive or unreacted dye might collate with DNA released from viable cells just prior to PCR and produce false-negative results.
Non-specific binding
High levels of dead and injured cells of either or both the target and non-target organism in the sample may mean that insufficient dye is available to target all dead cells leading to false-positive results.
Non-specific binding
High levels of dead and injured cells of either or both the target and non-target organism in the sample may mean that insufficient dye is available to target all dead cells leading to false-positive results.
Cell membrane integrity
Cell membrane composition of organisms naturally vary as a cell enters different growth phases. Membrane permeability of actively multiplying cells is thought to fluctuate as cell wall composition undergoes changes during active growth. Changes certainly occur during the stress created from the food manufacturing process. Environmental changes that cells encounter such as temperature or reduced moisture levels may lead to susceptibility to compounds such as EMA/ PMA. The presence of chemicals/detergents from cleaning and/or competition for nutrients may result in sub-lethally injured and starved cells. All of the conditions above may lead to living pathogenic cells absorbing the dyes and being killed, potentially leading to false-negative results.
Salt concentration and pH
Acidic conditions are known to reduce the performance of DNA intercalating dyes. Restoring or maintaining the sample at pH 5 can improve the performance of the dye. To adjust the pH of the sample prior to analysis may require a deviation from following standard reference methods and may not be an acceptable solution in a validated environment. Many food samples will contain quite high levels of salt to help increase their shelf-life. Salt concentration has been found to influence the binding of PMA which may preclude it from being used in these types of food matrices.
The above findings indicate the use of photoreactive DNA intercalating dyes might require control samples in conjunction with the test samples to monitor the effectiveness of the dyes when used in PCR testing of food samples. We suggest that changes in the Ct between PMA/EMA treated and untreated samples should be monitored to draw conclusions on the presence of dead cells.
In conclusion, it may be considered that the use of these dyes and chemicals does not always provide the solution expected. If they are incorporated into assays, it is important to fully understand the potential impact on results obtained. Even a protocol that is validated to use these reagents may not be able to overcome all the problem criteria for all food sample types. The possible advantages of using these reagents need careful consideration that takes account of the extra costs, the need for additional protocol steps, and the potential for reduced reliability of results.
Information courtesy of Thermo Scientific


